Subproject 3
Regulation of organohalide respiration in Dehalococcoides strains
Ute Lechner (University of Halle-Wittenberg)
The aim of this project is to characterize the mechanism of regulation of reductive dehalogenase (rdh) gene expression, which enables Dehalococcoides to respond specifically to diverse chloroorganic compounds and to perform multi-step dechlorination reactions. The role of MarR-type regulators and two-component-system regulators encoded in the genome by genes divergently transcribed to rdhA genes will be investigated. The work in this project will comprise
- transcription analysis of rdhA genes in response to specific chlorinated compounds
- identification of regulators/RdHs involved in the recognition and performance of multi-step reductive dechlorination reactions
- biochemical analysis of heterologously produced regulator proteins
- interaction of regulators with promoters of rdhA genes and possible ligands
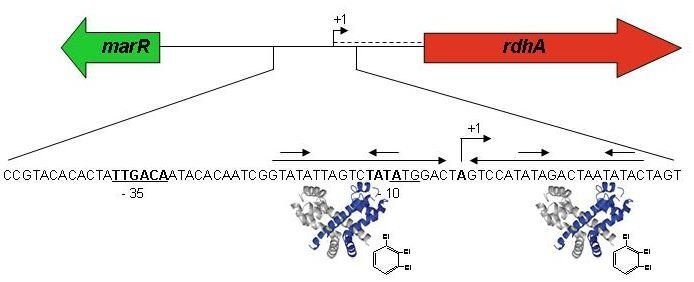
Model for the transcriptional control of rdhA gene expression by a MarR-type regulator interacting with a chlorinated compound
Selected References:
